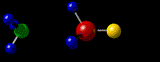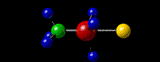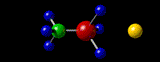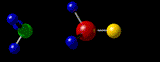
Containing a very brief, very selective and very summarized Yet Another Personal History Of The Web.
By the late nineties, the evolution of networks and (free) molecular visualization software allowed me to post on the world wide web, interactive, three-dimensional (3D) representations of molecular structures that I had “solved” or at least worked on, in my academic career twenty years earlier. My desire to do that has always been strong, simply because the pleasure I get from seeing those molecules in their 3D glory is huge, even though no-one else today would have the slightest interest. The problem for me has been preserving that capability over the twenty years since! Hence this 40-year-span post.
Origins
In the mid to late seventies for my Ph.D in chemical crystallography, I would send boxes of punched cards to London from Bristol University to calculate the three-dimensional structures of molecules on a CDC‑7600, a “super-computer” for the times. After about three weeks, if I hadn’t made a typing mistake with the instructions for the computer, the output would return in the mail in the form of a one-to-two inch thick printout from a line printer. (If I had made a mistake, I would have to correct the cards, resend the entire deck, and wait another few weeks). Then I would build a physical model of the molecule using colored balls (different colors for different atom types) on knitting needles stuck into a grid drawn on a polystyrene base, and at an angle, which together corresponded to the shape of the unit cell of the crystal. And the geometry of the molecular structure would be revealed in glorious, tactile 3D (and in color!)

 In the lab, synthetic chemists were doing new organometallic chemistry and would crystallize their reaction products, usually for the first time ever in the history of the universe. We crystallographers would put their best crystal in an X-Ray beam, measure the diffraction pattern, and use the data collected to calculate the crystal unit cell and ultimately, data permitting, the 3D chemical structure inside it. After a year or two, the results came back from London as a file over a network instead of a paper printout, and later, I can’t remember exactly when, we could send the input to London over a network too.
In the lab, synthetic chemists were doing new organometallic chemistry and would crystallize their reaction products, usually for the first time ever in the history of the universe. We crystallographers would put their best crystal in an X-Ray beam, measure the diffraction pattern, and use the data collected to calculate the crystal unit cell and ultimately, data permitting, the 3D chemical structure inside it. After a year or two, the results came back from London as a file over a network instead of a paper printout, and later, I can’t remember exactly when, we could send the input to London over a network too.
A decade later, in the mid-eighties at Edinburgh University I installed system software on a DEC VAX computer running VMS in the Department of Biochemistry to connect to academic networks JANET in the UK and ARPANET in the USA although UK domain names were still big-endian (uk.ac.edinburgh) while US domains were little-endian (dec.com) and the software had to convert between the two. In the end, little-endian prevailed and the UK had to do a 180º turn (towards edinburgh.ac.uk).
After another decade, in the mid-nineties, the Internet exploded into public and commercial use after Tim Berners-Lee invented HTML.
 In the early eighties we still also built large three-dimensional protein structures by hand, using a Richard’s box, half-silvered mirror, spotlights, brass atoms and plumb lines, for example. But as post-doc’s Phil Bourne and I used to drive up to Leeds University from Sheffield University in 1980 to use a new and evolving computer graphics system to build the protein structure “in the computer”. Molecular visualization software evolved enormously through the eighties and nineties supported by computer workstations for molecular graphics from the likes of Evans & Sutherland and Silicon Graphics. Five years after Sheffield and Leeds I used the VAX at Edinburgh to build and display the structures of biological molecules determined by X-Ray diffraction.
In the early eighties we still also built large three-dimensional protein structures by hand, using a Richard’s box, half-silvered mirror, spotlights, brass atoms and plumb lines, for example. But as post-doc’s Phil Bourne and I used to drive up to Leeds University from Sheffield University in 1980 to use a new and evolving computer graphics system to build the protein structure “in the computer”. Molecular visualization software evolved enormously through the eighties and nineties supported by computer workstations for molecular graphics from the likes of Evans & Sutherland and Silicon Graphics. Five years after Sheffield and Leeds I used the VAX at Edinburgh to build and display the structures of biological molecules determined by X-Ray diffraction.
The Internet
By the mid- to late-nineties (twenty years ago now) by which time I had moved with my family to the USA, I could hand-code a simple website at home and include some of the molecules I had worked on, in an interactive 3D representation, even if the interaction was limited to zooming, rotating and spinning, and stereo views. And for whatever reason, that satisfied a powerful desire to see “my” molecules in 3D again, even many years after I worked on them.
Chime
![]() I started building web pages manually, before the separation of form (with CSS) from content (with HTML) on home PCs running, first Windows NT Workstation 4.0, then Windows 95 then Windows ME then Windows 98 etc. One of the HTML editors I used was called CoffeeHTML and apparently still exists. An inventory of my software from 1998 also includes: Hot Dog Pro from Sausage Software and FlexED from InfoFlex for HTML editing; Wingif and LView Pro for image editing; WebLab Viewer from MSI and RASMOL for viewing molecules; and the Chime and Chime Pro plugins from MDL for viewing molecules in web pages. My first website containing interactive molecules was hand-built and depended on the Chime plugin. At some point between 1997 and 2004 I also started using Microsoft FrontPage for web publishing.
I started building web pages manually, before the separation of form (with CSS) from content (with HTML) on home PCs running, first Windows NT Workstation 4.0, then Windows 95 then Windows ME then Windows 98 etc. One of the HTML editors I used was called CoffeeHTML and apparently still exists. An inventory of my software from 1998 also includes: Hot Dog Pro from Sausage Software and FlexED from InfoFlex for HTML editing; Wingif and LView Pro for image editing; WebLab Viewer from MSI and RASMOL for viewing molecules; and the Chime and Chime Pro plugins from MDL for viewing molecules in web pages. My first website containing interactive molecules was hand-built and depended on the Chime plugin. At some point between 1997 and 2004 I also started using Microsoft FrontPage for web publishing.
Jmol
![]() By 2007 the proprietary Chime plugin had become obsolete and was being replaced by an open-source library called Jmol which used java applets to display molecules. Jmol became huge in education and display of 3d molecular structures in academic science. And by 2008 I was migrating my family to Apple Macintosh computers and phasing out Windows PC’s at home.
By 2007 the proprietary Chime plugin had become obsolete and was being replaced by an open-source library called Jmol which used java applets to display molecules. Jmol became huge in education and display of 3d molecular structures in academic science. And by 2008 I was migrating my family to Apple Macintosh computers and phasing out Windows PC’s at home.
In 2008 I started using Apple’s iWeb and built a first website for The Assembly of Geod (TAOG), my first large-scale steel geodesic sculpture. In 2012 I consolidated all three geodesic sculpture projects (TAOG, The Inaugural Balls, and The Dream Concentrator) into http://www.philomorph.net using iWeb, which is still a live site. But iWeb was a closed system (hey, it was from Apple) and I don’t believe I got Jmol working with iWeb.
But I was successful using a new program called RapidWeaver from Realmac Software. You just had to upload the Jmol files into the main folder for the website that was published by RapidWeaver, and make sure that each HTML page containing molecules made a call in its <head> to initialize the library.
From 2008 onwards, my molecules were spinning at https://robstansfield.site built with RapidWeaver and Jmol, but as you know, then java applets became the problem. Increasingly, my website just showed visitors the error message
You do not have Java applets enabled in your web browser, or your browser is blocking this applet. Check the warning message from your browser and/or enable Java applets in your web browser preferences, or install the Java Runtime Environment from www.java.com.
My molecules went into in hibernation for several years.
JSmol
 In the last few years I had read that JMol had been re-worked to use only HTML and CSS, but it was only just last week that I had a go at resuscitating my molecules. The Jmol library was converted to Javascript in about 2014 and works as promised. (Because of my unfamiliarity with Javascript it took me an entire day to understand how to display check boxes for such as “spin on/off” alongside a molecule. Duh!)
In the last few years I had read that JMol had been re-worked to use only HTML and CSS, but it was only just last week that I had a go at resuscitating my molecules. The Jmol library was converted to Javascript in about 2014 and works as promised. (Because of my unfamiliarity with Javascript it took me an entire day to understand how to display check boxes for such as “spin on/off” alongside a molecule. Duh!)
But I’m happy to report that I can spin molecules again, by adding JSmol (the Jmol Javascript Object) to a website published using RapidWeaver. So why are my molecules not spinning again on this site?
WordPress
 Because a month ago, I rebuilt this website using the (free) WordPress.org to support a blog, update the site in general, and to put my updated resume online. I was also curious about the pros and cons of WordPress as compared with RapidWeaver. But I more or less abandoned the molecules, or at least deliberately postponed their resuscitation.
Because a month ago, I rebuilt this website using the (free) WordPress.org to support a blog, update the site in general, and to put my updated resume online. I was also curious about the pros and cons of WordPress as compared with RapidWeaver. But I more or less abandoned the molecules, or at least deliberately postponed their resuscitation.
JSmol2wp
And then I discovered that Jim Hu, a computational biologist and one of the authors of the conversion of Jmol into JSmol, had released JSmol as a plugin for WordPress in early 2015! Well, I installed the plugin and it didn’t work. But when I inspected where the files went in the WordPress file structure, and copied over corresponding files from the most recent release of JSmol, it did work! It’s a little ugly yet, but if I can get to grips with the CSS for JSmol, I might be able to make it pretty enough to satisfy my own standards for visual acceptability.
MAMP
![]() This post was originally going to be about MAMP, but my website experiments enabled by MAMP Pro became much more interesting. Installing MAMP Pro on my laptop enabled me to create multiple test websites using WordPress and JSmol2wp, and RapidWeaver and JSmol, and to host them locally without mucking up a live site on the Internet. One of the most surprising features I found was the ability to “synchronize” via DropBox a host (the combination of web server and web content including MySQL databases) so that I could work on the same site from either of two different computers. Very cool.
This post was originally going to be about MAMP, but my website experiments enabled by MAMP Pro became much more interesting. Installing MAMP Pro on my laptop enabled me to create multiple test websites using WordPress and JSmol2wp, and RapidWeaver and JSmol, and to host them locally without mucking up a live site on the Internet. One of the most surprising features I found was the ability to “synchronize” via DropBox a host (the combination of web server and web content including MySQL databases) so that I could work on the same site from either of two different computers. Very cool.
The future
I’m not there yet, but either: I may be able to get my molecules spinning prettily within this site built with WordPress, or; I may be able to start from scratch with RapidWeaver and include both molecules and a blog to my taste. But from experience, either one could take years…
Postscript on Dec 2, 2017
A3 η2 Complexes of Cyclic Polyolefins: Crystal Structure of [Mn(CO)2(η2-C8H8)(η-C5H5)]. I.B.Benson, S.A.R.Knox, R.F.D.Stansfield and P.Woodward,
Journal of the Chemical Society, Dalton Transactions, (1981) 51-55.
[jsmol acc=’ix’ type=’mol’ wrap=6 commands=’=spin on’]
I’m not controlling the JSmol display using CSS, partly because JSmol2wp uses shortcodes (WordPress macros), but it works and the button styles in this theme are reasonable. Ta da!